用於分子特性預測的消息傳遞神經網路(MPNN)
作者: akensert
建立日期 2021/08/16
上次修改日期 2021/12/27
描述: 實現 MPNN 以預測血腦屏障的通透性。
簡介
在本教學中,我們將實作一種稱為消息傳遞神經網路 (MPNN) 的圖神經網路 (GNN),以預測圖的屬性。具體來說,我們將實作一個 MPNN 來預測一種稱為血腦屏障通透性 (BBBP) 的分子屬性。
動機:由於分子自然地以無向圖 G = (V, E) 表示,其中 V 是一組頂點(節點;原子),而 E 是一組邊(鍵),因此 GNN(例如 MPNN)正被證明是預測分子特性的有用方法。
到目前為止,更傳統的方法,例如隨機森林、支持向量機等,通常用於預測分子特性。與 GNN 相比,這些傳統方法通常基於預先計算的分子特徵進行操作,例如分子量、極性、電荷、碳原子數等。儘管這些分子特徵被證明是各種分子特性的良好預測因子,但假設基於這些更「原始」、「低階」的特徵進行操作可能會更好。
參考文獻
近年來,人們投入了大量精力來開發用於圖形數據(包括分子圖形)的神經網路。有關圖神經網路的摘要,請參閱 圖神經網路的綜合調查 和 圖神經網路:方法和應用綜述;若要進一步閱讀本教學中實作的特定圖神經網路,請參閱 量子化學的神經訊息傳遞 和 DeepChem 的 MPNNModel。
設定
安裝 RDKit 和其他依賴項
(以下文字取自此教學)。
RDKit 是一個以 C++ 和 Python 編寫的化學資訊學和機器學習軟體集合。在本教學中,RDKit 用於方便有效地將 SMILES 轉換為分子物件,然後從這些物件取得原子和鍵的集合。
SMILES 以 ASCII 字串的形式表示給定分子的結構。SMILES 字串是一種緊湊的編碼,對於較小的分子,它具有相對的人類可讀性。將分子編碼為字串既減輕了給定分子的資料庫和/或網路搜尋的負擔,也使其變得更容易。RDKit 使用演算法精確地將給定的 SMILES 轉換為分子物件,然後該物件可用於計算大量分子特性/特徵。
請注意,RDKit 通常透過 Conda 安裝。然而,由於 rdkit_platform_wheels,rdkit 現在可以(為了本教學的目的)透過 pip 輕鬆安裝,如下所示
pip -q install rdkit-pypi
為了方便有效地讀取 csv 檔案和視覺化,需要安裝以下內容
pip -q install pandas
pip -q install Pillow
pip -q install matplotlib
pip -q install pydot
sudo apt-get -qq install graphviz
匯入套件
import os
# Temporary suppress tf logs
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import warnings
from rdkit import Chem
from rdkit import RDLogger
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem.Draw import MolsToGridImage
# Temporary suppress warnings and RDKit logs
warnings.filterwarnings("ignore")
RDLogger.DisableLog("rdApp.*")
np.random.seed(42)
tf.random.set_seed(42)
資料集
有關資料集的資訊,請參閱 矽基血腦屏障穿透建模的貝葉斯方法 和 MoleculeNet:分子機器學習的基準。該資料集將從 MoleculeNet.org 下載。
關於
該資料集包含 2,050 個分子。每個分子都帶有名稱、標籤和 SMILES 字串。
血腦屏障 (BBB) 是一種將血液與腦細胞外液分離的膜,因此會阻止大多數藥物(分子)到達大腦。因此,BBBP 對於開發旨在針對中樞神經系統的新藥非常重要。此資料集的標籤是二元的(1 或 0),並表示分子的通透性。
csv_path = keras.utils.get_file(
"BBBP.csv", "https://deepchemdata.s3-us-west-1.amazonaws.com/datasets/BBBP.csv"
)
df = pd.read_csv(csv_path, usecols=[1, 2, 3])
df.iloc[96:104]
| 名稱 | p_np | smiles | |
|---|---|---|---|
| 96 | 頭孢西丁 | 1 | CO[C@]1(NC(=O)Cc2sccc2)[C@H]3SCC(=C(N3C1=O)C(O... |
| 97 | Org34167 | 1 | NC(CC=C)c1ccccc1c2noc3c2cccc3 |
| 98 | 9-OH 利培酮 | 1 | OC1C(N2CCC1)=NC(C)=C(CCN3CCC(CC3)c4c5ccc(F)cc5... |
| 99 | 對乙醯氨基酚 | 1 | CC(=O)Nc1ccc(O)cc1 |
| 100 | 乙醯水楊酸鹽 | 0 | CC(=O)Oc1ccccc1C(O)=O |
| 101 | 別嘌呤醇 | 0 | O=C1N=CN=C2NNC=C12 |
| 102 | 前列地爾 | 0 | CCCCC[C@H](O)/C=C/[C@H]1[C@H](O)CC(=O)[C@@H]1C... |
| 103 | 氨茶鹼 | 0 | CN1C(=O)N(C)c2nc[nH]c2C1=O.CN3C(=O)N(C)c4nc[nH... |
定義特徵
為了編碼原子和鍵的特徵(稍後我們將需要這些特徵),我們將定義兩個類別:分別是 AtomFeaturizer 和 BondFeaturizer。
為了減少程式碼行數,即讓本教學簡短精煉,只會考慮少數(原子和鍵)特徵:[原子特徵] 符號(元素)、價電子數、氫鍵數、軌道雜化、[鍵特徵] (共價)鍵類型和 共軛。
class Featurizer:
def __init__(self, allowable_sets):
self.dim = 0
self.features_mapping = {}
for k, s in allowable_sets.items():
s = sorted(list(s))
self.features_mapping[k] = dict(zip(s, range(self.dim, len(s) + self.dim)))
self.dim += len(s)
def encode(self, inputs):
output = np.zeros((self.dim,))
for name_feature, feature_mapping in self.features_mapping.items():
feature = getattr(self, name_feature)(inputs)
if feature not in feature_mapping:
continue
output[feature_mapping[feature]] = 1.0
return output
class AtomFeaturizer(Featurizer):
def __init__(self, allowable_sets):
super().__init__(allowable_sets)
def symbol(self, atom):
return atom.GetSymbol()
def n_valence(self, atom):
return atom.GetTotalValence()
def n_hydrogens(self, atom):
return atom.GetTotalNumHs()
def hybridization(self, atom):
return atom.GetHybridization().name.lower()
class BondFeaturizer(Featurizer):
def __init__(self, allowable_sets):
super().__init__(allowable_sets)
self.dim += 1
def encode(self, bond):
output = np.zeros((self.dim,))
if bond is None:
output[-1] = 1.0
return output
output = super().encode(bond)
return output
def bond_type(self, bond):
return bond.GetBondType().name.lower()
def conjugated(self, bond):
return bond.GetIsConjugated()
atom_featurizer = AtomFeaturizer(
allowable_sets={
"symbol": {"B", "Br", "C", "Ca", "Cl", "F", "H", "I", "N", "Na", "O", "P", "S"},
"n_valence": {0, 1, 2, 3, 4, 5, 6},
"n_hydrogens": {0, 1, 2, 3, 4},
"hybridization": {"s", "sp", "sp2", "sp3"},
}
)
bond_featurizer = BondFeaturizer(
allowable_sets={
"bond_type": {"single", "double", "triple", "aromatic"},
"conjugated": {True, False},
}
)
產生圖形
在我們可以從 SMILES 產生完整圖形之前,我們需要實作以下函數
-
molecule_from_smiles,它將 SMILES 作為輸入並傳回分子物件。這一切都由 RDKit 處理。 -
graph_from_molecule,它將分子物件作為輸入並傳回一個圖,該圖表示為三元組 (atom_features, bond_features, pair_indices)。為此,我們將使用先前定義的類別。
最後,我們現在可以實作函數 graphs_from_smiles,它在訓練、驗證和測試資料集的所有 SMILES 上應用函數 (1) 和隨後的 (2)。
注意:雖然建議對此資料集進行支架分割(請參閱 此處),但為了簡單起見,我們執行了簡單的隨機分割。
def molecule_from_smiles(smiles):
# MolFromSmiles(m, sanitize=True) should be equivalent to
# MolFromSmiles(m, sanitize=False) -> SanitizeMol(m) -> AssignStereochemistry(m, ...)
molecule = Chem.MolFromSmiles(smiles, sanitize=False)
# If sanitization is unsuccessful, catch the error, and try again without
# the sanitization step that caused the error
flag = Chem.SanitizeMol(molecule, catchErrors=True)
if flag != Chem.SanitizeFlags.SANITIZE_NONE:
Chem.SanitizeMol(molecule, sanitizeOps=Chem.SanitizeFlags.SANITIZE_ALL ^ flag)
Chem.AssignStereochemistry(molecule, cleanIt=True, force=True)
return molecule
def graph_from_molecule(molecule):
# Initialize graph
atom_features = []
bond_features = []
pair_indices = []
for atom in molecule.GetAtoms():
atom_features.append(atom_featurizer.encode(atom))
# Add self-loops
pair_indices.append([atom.GetIdx(), atom.GetIdx()])
bond_features.append(bond_featurizer.encode(None))
for neighbor in atom.GetNeighbors():
bond = molecule.GetBondBetweenAtoms(atom.GetIdx(), neighbor.GetIdx())
pair_indices.append([atom.GetIdx(), neighbor.GetIdx()])
bond_features.append(bond_featurizer.encode(bond))
return np.array(atom_features), np.array(bond_features), np.array(pair_indices)
def graphs_from_smiles(smiles_list):
# Initialize graphs
atom_features_list = []
bond_features_list = []
pair_indices_list = []
for smiles in smiles_list:
molecule = molecule_from_smiles(smiles)
atom_features, bond_features, pair_indices = graph_from_molecule(molecule)
atom_features_list.append(atom_features)
bond_features_list.append(bond_features)
pair_indices_list.append(pair_indices)
# Convert lists to ragged tensors for tf.data.Dataset later on
return (
tf.ragged.constant(atom_features_list, dtype=tf.float32),
tf.ragged.constant(bond_features_list, dtype=tf.float32),
tf.ragged.constant(pair_indices_list, dtype=tf.int64),
)
# Shuffle array of indices ranging from 0 to 2049
permuted_indices = np.random.permutation(np.arange(df.shape[0]))
# Train set: 80 % of data
train_index = permuted_indices[: int(df.shape[0] * 0.8)]
x_train = graphs_from_smiles(df.iloc[train_index].smiles)
y_train = df.iloc[train_index].p_np
# Valid set: 19 % of data
valid_index = permuted_indices[int(df.shape[0] * 0.8) : int(df.shape[0] * 0.99)]
x_valid = graphs_from_smiles(df.iloc[valid_index].smiles)
y_valid = df.iloc[valid_index].p_np
# Test set: 1 % of data
test_index = permuted_indices[int(df.shape[0] * 0.99) :]
x_test = graphs_from_smiles(df.iloc[test_index].smiles)
y_test = df.iloc[test_index].p_np
測試函數
print(f"Name:\t{df.name[100]}\nSMILES:\t{df.smiles[100]}\nBBBP:\t{df.p_np[100]}")
molecule = molecule_from_smiles(df.iloc[100].smiles)
print("Molecule:")
molecule
Name: acetylsalicylate
SMILES: CC(=O)Oc1ccccc1C(O)=O
BBBP: 0
Molecule:
graph = graph_from_molecule(molecule)
print("Graph (including self-loops):")
print("\tatom features\t", graph[0].shape)
print("\tbond features\t", graph[1].shape)
print("\tpair indices\t", graph[2].shape)
Graph (including self-loops):
atom features (13, 29)
bond features (39, 7)
pair indices (39, 2)
建立 tf.data.Dataset
在本教學中,MPNN 實作將(每次迭代)將單個圖作為輸入。因此,給定一批(子)圖形(分子),我們需要將它們合併到一個單個圖形中(我們將此圖形稱為全域圖形)。這個全域圖形是一個斷開的圖形,其中每個子圖形與其他子圖形完全分離。
def prepare_batch(x_batch, y_batch):
"""Merges (sub)graphs of batch into a single global (disconnected) graph
"""
atom_features, bond_features, pair_indices = x_batch
# Obtain number of atoms and bonds for each graph (molecule)
num_atoms = atom_features.row_lengths()
num_bonds = bond_features.row_lengths()
# Obtain partition indices (molecule_indicator), which will be used to
# gather (sub)graphs from global graph in model later on
molecule_indices = tf.range(len(num_atoms))
molecule_indicator = tf.repeat(molecule_indices, num_atoms)
# Merge (sub)graphs into a global (disconnected) graph. Adding 'increment' to
# 'pair_indices' (and merging ragged tensors) actualizes the global graph
gather_indices = tf.repeat(molecule_indices[:-1], num_bonds[1:])
increment = tf.cumsum(num_atoms[:-1])
increment = tf.pad(tf.gather(increment, gather_indices), [(num_bonds[0], 0)])
pair_indices = pair_indices.merge_dims(outer_axis=0, inner_axis=1).to_tensor()
pair_indices = pair_indices + increment[:, tf.newaxis]
atom_features = atom_features.merge_dims(outer_axis=0, inner_axis=1).to_tensor()
bond_features = bond_features.merge_dims(outer_axis=0, inner_axis=1).to_tensor()
return (atom_features, bond_features, pair_indices, molecule_indicator), y_batch
def MPNNDataset(X, y, batch_size=32, shuffle=False):
dataset = tf.data.Dataset.from_tensor_slices((X, (y)))
if shuffle:
dataset = dataset.shuffle(1024)
return dataset.batch(batch_size).map(prepare_batch, -1).prefetch(-1)
模型
MPNN 模型可以採用各種形狀和形式。在本教學中,我們將基於原始論文 量子化學的神經訊息傳遞 和 DeepChem 的 MPNNModel 實作 MPNN。本教學的 MPNN 包括三個階段:消息傳遞、讀取和分類。
消息傳遞
消息傳遞步驟本身包含兩個部分
-
邊緣網路,它根據它們之間的邊緣特徵 (
e_{vw_{i}}) 將v的 1-跳鄰居w_{i}的訊息傳遞到v,從而產生更新的節點(狀態)v'。w_{i}表示v的第i個鄰居。 -
閘控循環單元 (GRU),它將最新的節點狀態作為輸入,並根據先前的節點狀態更新它。換句話說,最新的節點狀態充當 GRU 的輸入,而先前的節點狀態則合併在 GRU 的記憶體狀態中。這允許資訊從一個節點狀態(例如,
v)傳遞到另一個節點狀態(例如,v'')。
重要的是,步驟 (1) 和 (2) 會重複 k 個步驟,並且在每個步驟 1...k 時,從 v 聚合的資訊的半徑(或跳數)增加 1。
class EdgeNetwork(layers.Layer):
def build(self, input_shape):
self.atom_dim = input_shape[0][-1]
self.bond_dim = input_shape[1][-1]
self.kernel = self.add_weight(
shape=(self.bond_dim, self.atom_dim * self.atom_dim),
initializer="glorot_uniform",
name="kernel",
)
self.bias = self.add_weight(
shape=(self.atom_dim * self.atom_dim), initializer="zeros", name="bias",
)
self.built = True
def call(self, inputs):
atom_features, bond_features, pair_indices = inputs
# Apply linear transformation to bond features
bond_features = tf.matmul(bond_features, self.kernel) + self.bias
# Reshape for neighborhood aggregation later
bond_features = tf.reshape(bond_features, (-1, self.atom_dim, self.atom_dim))
# Obtain atom features of neighbors
atom_features_neighbors = tf.gather(atom_features, pair_indices[:, 1])
atom_features_neighbors = tf.expand_dims(atom_features_neighbors, axis=-1)
# Apply neighborhood aggregation
transformed_features = tf.matmul(bond_features, atom_features_neighbors)
transformed_features = tf.squeeze(transformed_features, axis=-1)
aggregated_features = tf.math.unsorted_segment_sum(
transformed_features,
pair_indices[:, 0],
num_segments=tf.shape(atom_features)[0],
)
return aggregated_features
class MessagePassing(layers.Layer):
def __init__(self, units, steps=4, **kwargs):
super().__init__(**kwargs)
self.units = units
self.steps = steps
def build(self, input_shape):
self.atom_dim = input_shape[0][-1]
self.message_step = EdgeNetwork()
self.pad_length = max(0, self.units - self.atom_dim)
self.update_step = layers.GRUCell(self.atom_dim + self.pad_length)
self.built = True
def call(self, inputs):
atom_features, bond_features, pair_indices = inputs
# Pad atom features if number of desired units exceeds atom_features dim.
# Alternatively, a dense layer could be used here.
atom_features_updated = tf.pad(atom_features, [(0, 0), (0, self.pad_length)])
# Perform a number of steps of message passing
for i in range(self.steps):
# Aggregate information from neighbors
atom_features_aggregated = self.message_step(
[atom_features_updated, bond_features, pair_indices]
)
# Update node state via a step of GRU
atom_features_updated, _ = self.update_step(
atom_features_aggregated, atom_features_updated
)
return atom_features_updated
讀取
當消息傳遞過程結束時,k 步聚合的節點狀態將被劃分為子圖(對應於批次中的每個分子),然後縮減為圖級嵌入。在 原始論文 中,一個 設定為設定層 用於此目的。但在本教學中,將使用轉換器編碼器 + 平均池化。具體而言
- k 步聚合的節點狀態將被劃分為子圖(對應於批次中的每個分子);
- 然後,每個子圖將被填充以符合具有最大節點數的子圖,然後是
tf.stack(...); - (堆疊填充的)張量,編碼子圖(每個子圖包含一組節點狀態),會被遮罩,以確保填充不會干擾訓練;
- 最後,張量將傳遞到轉換器,然後進行平均池化。
class PartitionPadding(layers.Layer):
def __init__(self, batch_size, **kwargs):
super().__init__(**kwargs)
self.batch_size = batch_size
def call(self, inputs):
atom_features, molecule_indicator = inputs
# Obtain subgraphs
atom_features_partitioned = tf.dynamic_partition(
atom_features, molecule_indicator, self.batch_size
)
# Pad and stack subgraphs
num_atoms = [tf.shape(f)[0] for f in atom_features_partitioned]
max_num_atoms = tf.reduce_max(num_atoms)
atom_features_stacked = tf.stack(
[
tf.pad(f, [(0, max_num_atoms - n), (0, 0)])
for f, n in zip(atom_features_partitioned, num_atoms)
],
axis=0,
)
# Remove empty subgraphs (usually for last batch in dataset)
gather_indices = tf.where(tf.reduce_sum(atom_features_stacked, (1, 2)) != 0)
gather_indices = tf.squeeze(gather_indices, axis=-1)
return tf.gather(atom_features_stacked, gather_indices, axis=0)
class TransformerEncoderReadout(layers.Layer):
def __init__(
self, num_heads=8, embed_dim=64, dense_dim=512, batch_size=32, **kwargs
):
super().__init__(**kwargs)
self.partition_padding = PartitionPadding(batch_size)
self.attention = layers.MultiHeadAttention(num_heads, embed_dim)
self.dense_proj = keras.Sequential(
[layers.Dense(dense_dim, activation="relu"), layers.Dense(embed_dim),]
)
self.layernorm_1 = layers.LayerNormalization()
self.layernorm_2 = layers.LayerNormalization()
self.average_pooling = layers.GlobalAveragePooling1D()
def call(self, inputs):
x = self.partition_padding(inputs)
padding_mask = tf.reduce_any(tf.not_equal(x, 0.0), axis=-1)
padding_mask = padding_mask[:, tf.newaxis, tf.newaxis, :]
attention_output = self.attention(x, x, attention_mask=padding_mask)
proj_input = self.layernorm_1(x + attention_output)
proj_output = self.layernorm_2(proj_input + self.dense_proj(proj_input))
return self.average_pooling(proj_output)
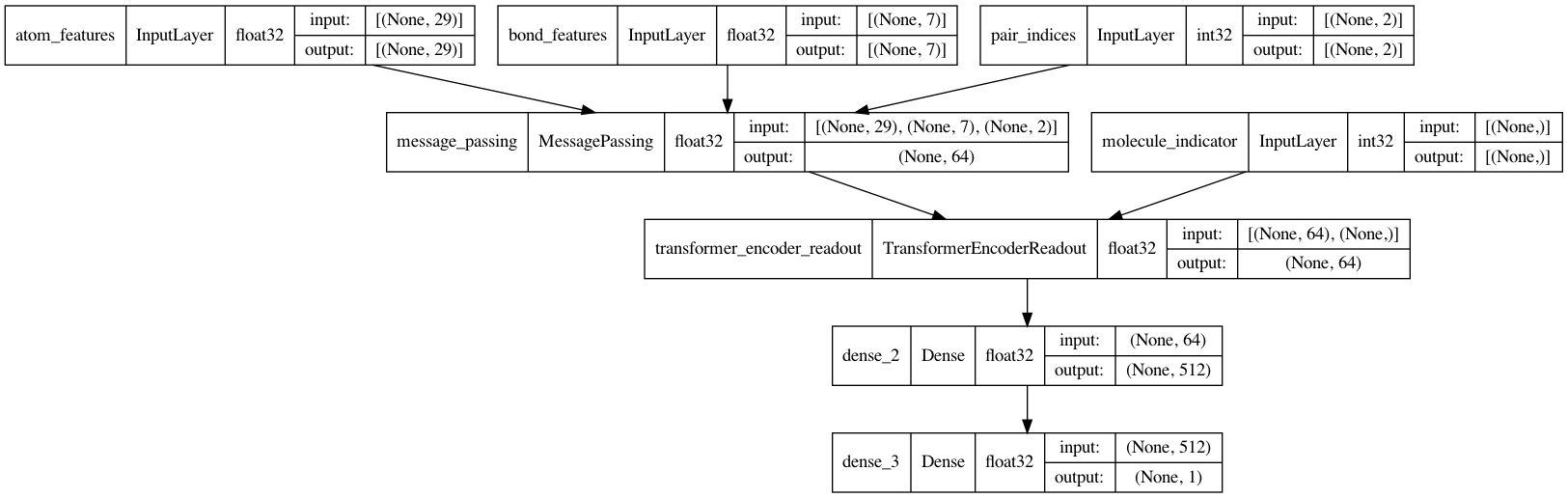
訊息傳遞神經網路(MPNN)
現在是時候完成 MPNN 模型了。除了訊息傳遞和讀取之外,還將實作一個兩層分類網路來預測 BBBP。
def MPNNModel(
atom_dim,
bond_dim,
batch_size=32,
message_units=64,
message_steps=4,
num_attention_heads=8,
dense_units=512,
):
atom_features = layers.Input((atom_dim), dtype="float32", name="atom_features")
bond_features = layers.Input((bond_dim), dtype="float32", name="bond_features")
pair_indices = layers.Input((2), dtype="int32", name="pair_indices")
molecule_indicator = layers.Input((), dtype="int32", name="molecule_indicator")
x = MessagePassing(message_units, message_steps)(
[atom_features, bond_features, pair_indices]
)
x = TransformerEncoderReadout(
num_attention_heads, message_units, dense_units, batch_size
)([x, molecule_indicator])
x = layers.Dense(dense_units, activation="relu")(x)
x = layers.Dense(1, activation="sigmoid")(x)
model = keras.Model(
inputs=[atom_features, bond_features, pair_indices, molecule_indicator],
outputs=[x],
)
return model
mpnn = MPNNModel(
atom_dim=x_train[0][0][0].shape[0], bond_dim=x_train[1][0][0].shape[0],
)
mpnn.compile(
loss=keras.losses.BinaryCrossentropy(),
optimizer=keras.optimizers.Adam(learning_rate=5e-4),
metrics=[keras.metrics.AUC(name="AUC")],
)
keras.utils.plot_model(mpnn, show_dtype=True, show_shapes=True)
訓練
train_dataset = MPNNDataset(x_train, y_train)
valid_dataset = MPNNDataset(x_valid, y_valid)
test_dataset = MPNNDataset(x_test, y_test)
history = mpnn.fit(
train_dataset,
validation_data=valid_dataset,
epochs=40,
verbose=2,
class_weight={0: 2.0, 1: 0.5},
)
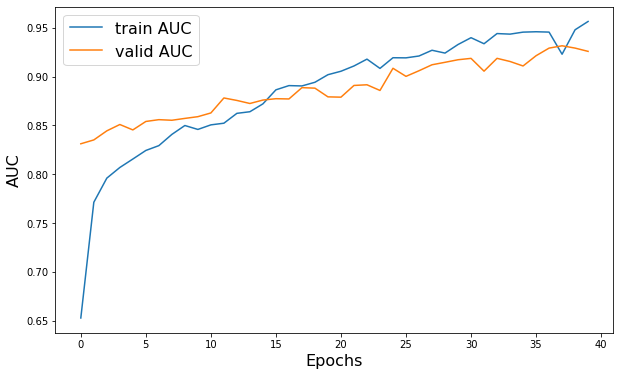
plt.figure(figsize=(10, 6))
plt.plot(history.history["AUC"], label="train AUC")
plt.plot(history.history["val_AUC"], label="valid AUC")
plt.xlabel("Epochs", fontsize=16)
plt.ylabel("AUC", fontsize=16)
plt.legend(fontsize=16)
Epoch 1/40
52/52 - 26s - loss: 0.5572 - AUC: 0.6527 - val_loss: 0.4660 - val_AUC: 0.8312 - 26s/epoch - 501ms/step
Epoch 2/40
52/52 - 22s - loss: 0.4817 - AUC: 0.7713 - val_loss: 0.6889 - val_AUC: 0.8351 - 22s/epoch - 416ms/step
Epoch 3/40
52/52 - 24s - loss: 0.4611 - AUC: 0.7960 - val_loss: 0.5863 - val_AUC: 0.8444 - 24s/epoch - 457ms/step
Epoch 4/40
52/52 - 19s - loss: 0.4493 - AUC: 0.8069 - val_loss: 0.5059 - val_AUC: 0.8509 - 19s/epoch - 372ms/step
Epoch 5/40
52/52 - 21s - loss: 0.4420 - AUC: 0.8155 - val_loss: 0.4965 - val_AUC: 0.8454 - 21s/epoch - 405ms/step
Epoch 6/40
52/52 - 22s - loss: 0.4344 - AUC: 0.8243 - val_loss: 0.5307 - val_AUC: 0.8540 - 22s/epoch - 419ms/step
Epoch 7/40
52/52 - 26s - loss: 0.4301 - AUC: 0.8293 - val_loss: 0.5131 - val_AUC: 0.8559 - 26s/epoch - 503ms/step
Epoch 8/40
52/52 - 31s - loss: 0.4163 - AUC: 0.8408 - val_loss: 0.5361 - val_AUC: 0.8552 - 31s/epoch - 599ms/step
Epoch 9/40
52/52 - 30s - loss: 0.4095 - AUC: 0.8499 - val_loss: 0.5371 - val_AUC: 0.8572 - 30s/epoch - 578ms/step
Epoch 10/40
52/52 - 23s - loss: 0.4107 - AUC: 0.8459 - val_loss: 0.5923 - val_AUC: 0.8589 - 23s/epoch - 444ms/step
Epoch 11/40
52/52 - 29s - loss: 0.4107 - AUC: 0.8505 - val_loss: 0.5070 - val_AUC: 0.8627 - 29s/epoch - 553ms/step
Epoch 12/40
52/52 - 25s - loss: 0.4005 - AUC: 0.8522 - val_loss: 0.5417 - val_AUC: 0.8781 - 25s/epoch - 471ms/step
Epoch 13/40
52/52 - 22s - loss: 0.3924 - AUC: 0.8623 - val_loss: 0.5915 - val_AUC: 0.8755 - 22s/epoch - 425ms/step
Epoch 14/40
52/52 - 19s - loss: 0.3872 - AUC: 0.8640 - val_loss: 0.5852 - val_AUC: 0.8724 - 19s/epoch - 365ms/step
Epoch 15/40
52/52 - 19s - loss: 0.3812 - AUC: 0.8720 - val_loss: 0.4949 - val_AUC: 0.8759 - 19s/epoch - 362ms/step
Epoch 16/40
52/52 - 27s - loss: 0.3604 - AUC: 0.8864 - val_loss: 0.5076 - val_AUC: 0.8773 - 27s/epoch - 521ms/step
Epoch 17/40
52/52 - 37s - loss: 0.3554 - AUC: 0.8907 - val_loss: 0.4556 - val_AUC: 0.8771 - 37s/epoch - 712ms/step
Epoch 18/40
52/52 - 23s - loss: 0.3554 - AUC: 0.8904 - val_loss: 0.4854 - val_AUC: 0.8887 - 23s/epoch - 452ms/step
Epoch 19/40
52/52 - 26s - loss: 0.3504 - AUC: 0.8942 - val_loss: 0.4622 - val_AUC: 0.8881 - 26s/epoch - 507ms/step
Epoch 20/40
52/52 - 20s - loss: 0.3378 - AUC: 0.9019 - val_loss: 0.5568 - val_AUC: 0.8792 - 20s/epoch - 390ms/step
Epoch 21/40
52/52 - 19s - loss: 0.3324 - AUC: 0.9055 - val_loss: 0.5623 - val_AUC: 0.8789 - 19s/epoch - 363ms/step
Epoch 22/40
52/52 - 19s - loss: 0.3248 - AUC: 0.9109 - val_loss: 0.5486 - val_AUC: 0.8909 - 19s/epoch - 357ms/step
Epoch 23/40
52/52 - 18s - loss: 0.3126 - AUC: 0.9179 - val_loss: 0.5684 - val_AUC: 0.8916 - 18s/epoch - 348ms/step
Epoch 24/40
52/52 - 18s - loss: 0.3296 - AUC: 0.9084 - val_loss: 0.5462 - val_AUC: 0.8858 - 18s/epoch - 352ms/step
Epoch 25/40
52/52 - 18s - loss: 0.3098 - AUC: 0.9193 - val_loss: 0.4212 - val_AUC: 0.9085 - 18s/epoch - 349ms/step
Epoch 26/40
52/52 - 18s - loss: 0.3095 - AUC: 0.9192 - val_loss: 0.4991 - val_AUC: 0.9002 - 18s/epoch - 348ms/step
Epoch 27/40
52/52 - 18s - loss: 0.3056 - AUC: 0.9211 - val_loss: 0.4739 - val_AUC: 0.9060 - 18s/epoch - 349ms/step
Epoch 28/40
52/52 - 18s - loss: 0.2942 - AUC: 0.9270 - val_loss: 0.4188 - val_AUC: 0.9121 - 18s/epoch - 344ms/step
Epoch 29/40
52/52 - 18s - loss: 0.3004 - AUC: 0.9241 - val_loss: 0.4056 - val_AUC: 0.9146 - 18s/epoch - 351ms/step
Epoch 30/40
52/52 - 18s - loss: 0.2810 - AUC: 0.9328 - val_loss: 0.3923 - val_AUC: 0.9172 - 18s/epoch - 355ms/step
Epoch 31/40
52/52 - 18s - loss: 0.2661 - AUC: 0.9398 - val_loss: 0.3609 - val_AUC: 0.9186 - 18s/epoch - 349ms/step
Epoch 32/40
52/52 - 19s - loss: 0.2797 - AUC: 0.9336 - val_loss: 0.3764 - val_AUC: 0.9055 - 19s/epoch - 357ms/step
Epoch 33/40
52/52 - 19s - loss: 0.2552 - AUC: 0.9441 - val_loss: 0.3941 - val_AUC: 0.9187 - 19s/epoch - 368ms/step
Epoch 34/40
52/52 - 23s - loss: 0.2601 - AUC: 0.9435 - val_loss: 0.4128 - val_AUC: 0.9154 - 23s/epoch - 443ms/step
Epoch 35/40
52/52 - 32s - loss: 0.2533 - AUC: 0.9455 - val_loss: 0.4191 - val_AUC: 0.9109 - 32s/epoch - 615ms/step
Epoch 36/40
52/52 - 23s - loss: 0.2530 - AUC: 0.9459 - val_loss: 0.4276 - val_AUC: 0.9213 - 23s/epoch - 435ms/step
Epoch 37/40
52/52 - 31s - loss: 0.2531 - AUC: 0.9456 - val_loss: 0.3950 - val_AUC: 0.9292 - 31s/epoch - 593ms/step
Epoch 38/40
52/52 - 22s - loss: 0.3039 - AUC: 0.9229 - val_loss: 0.3114 - val_AUC: 0.9315 - 22s/epoch - 428ms/step
Epoch 39/40
52/52 - 20s - loss: 0.2477 - AUC: 0.9479 - val_loss: 0.3584 - val_AUC: 0.9292 - 20s/epoch - 391ms/step
Epoch 40/40
52/52 - 22s - loss: 0.2276 - AUC: 0.9565 - val_loss: 0.3279 - val_AUC: 0.9258 - 22s/epoch - 416ms/step
<matplotlib.legend.Legend at 0x1603c63d0>
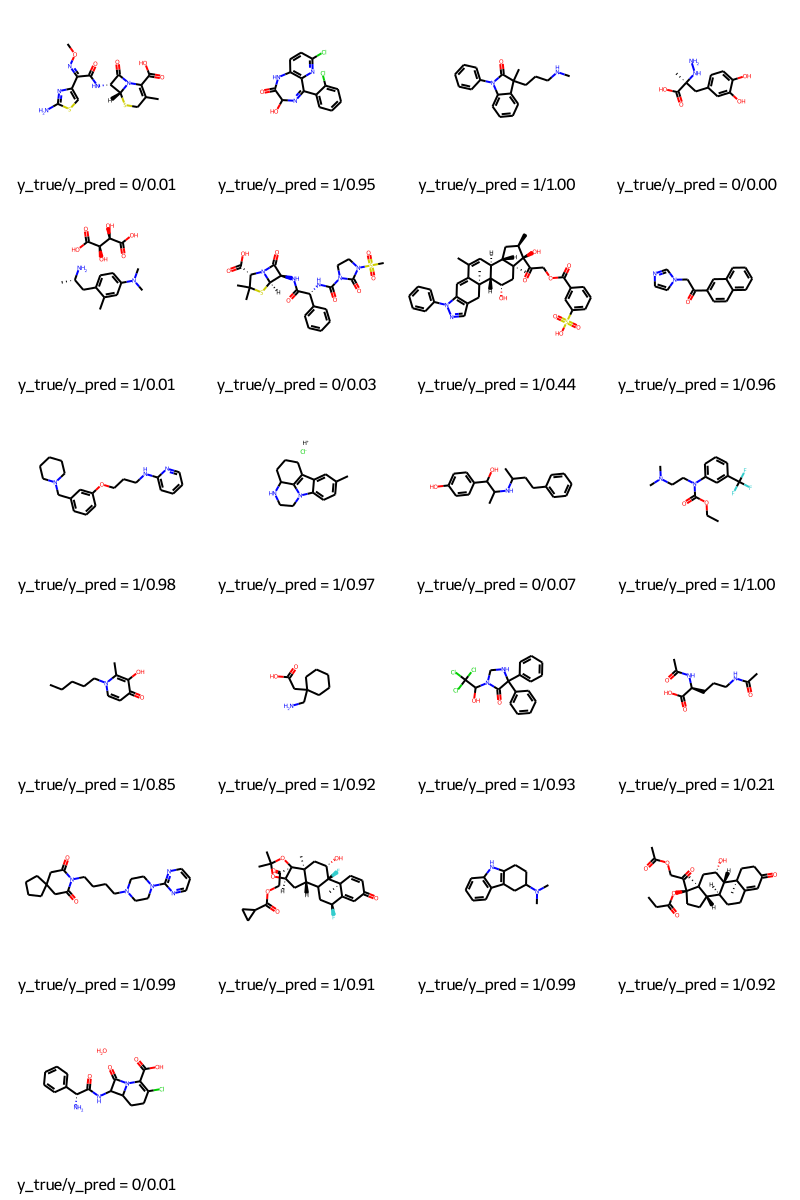
預測
molecules = [molecule_from_smiles(df.smiles.values[index]) for index in test_index]
y_true = [df.p_np.values[index] for index in test_index]
y_pred = tf.squeeze(mpnn.predict(test_dataset), axis=1)
legends = [f"y_true/y_pred = {y_true[i]}/{y_pred[i]:.2f}" for i in range(len(y_true))]
MolsToGridImage(molecules, molsPerRow=4, legends=legends)
結論
在本教學中,我們示範了一個訊息傳遞神經網路 (MPNN),以預測許多不同分子的血腦屏障通透性 (BBBP)。我們首先必須從 SMILES 建構圖形,然後建立一個可以在這些圖形上運作的 Keras 模型,最後訓練該模型以進行預測。
HuggingFace 上提供的範例
| 訓練模型 | 示範 |
|---|---|
 |
 |
